Click on the image to see a PDF version (for zooming in)
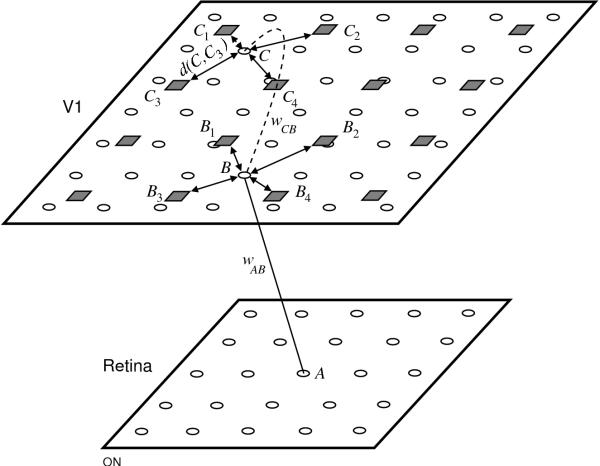
Fig. 15.5. Weight interpolation in GLISSOM. This example shows
a V1 of size 4 × 4 being scaled to 7 × 7, with a fixed 8 × 8
retina. Both V1 networks are plotted in a continuous two-dimensional
area representing the surface of the cortex. The squares in V1
represent neurons in the original network (i.e. before scaling) and
circles represent neurons in the new, scaled network. A is a retinal
receptor cell and B and C are neurons in the new network. Afferent
connection strengths to neuron B in the new network are calculated
based on the connection strengths of the ancestors of B, i.e. those
neurons in the original network that surround the position of B
(B1, B2, B3, and B4 in
this case). The new afferent connection strength wAB from
receptor A to B is a normalized combination of the connection
strengths wABi from A to each ancestor
Bi of B, weighted inversely by the distance d(B,
Bi) between Bi and B. Lateral connection
strengths from C to B are calculated similarly, as a
proximity-weighted combination of the connection strengths between the
ancestors of those neurons. Thus, the connection strengths in the
scaled network consist of proximity-weighted combinations of the
connection strengths in the original network.
|