Logic Techniques Applied to Genomics
 The progression of cancer (or any genetically rooted disease) goes through a
sequence of non-cancerous genetic states, which terminate in a cycle of cancerous
genetic states (also referred to as an attractor cycle). Biologists have identified
these attractor states in cancerous patients through laboratory experiments. These
sequences of states can be modeled as a Finite State Machine (FSM), and a slew of
design and analysis techniques in the area of Logic Synthesis can be used
to understand and model the problem.
However, unlike an FSM design
problem in VLSI, the FSM for cancer is not known, and has to be determined. In VLSI
design, we first design the FSM, and then reason about its reachable states,
terminal (attractor) cycles etc. In the context of cancer, the FSM (also called the
gene regulatory network or GRN) has to be derived from observed data
corresponding to the genetic states that are visited in an attractor cycle.
The progression of cancer (or any genetically rooted disease) goes through a
sequence of non-cancerous genetic states, which terminate in a cycle of cancerous
genetic states (also referred to as an attractor cycle). Biologists have identified
these attractor states in cancerous patients through laboratory experiments. These
sequences of states can be modeled as a Finite State Machine (FSM), and a slew of
design and analysis techniques in the area of Logic Synthesis can be used
to understand and model the problem.
However, unlike an FSM design
problem in VLSI, the FSM for cancer is not known, and has to be determined. In VLSI
design, we first design the FSM, and then reason about its reachable states,
terminal (attractor) cycles etc. In the context of cancer, the FSM (also called the
gene regulatory network or GRN) has to be derived from observed data
corresponding to the genetic states that are visited in an attractor cycle.
We have developed a powerful Boolean Satisfiability (SAT) based approach to infer
the genes that regulate a particular gene in the GRN
Using this information, we are now working on techniques to
develop the FSM (GRN) in a cancerous patient, using GPUs to accelerate the
computation. Previous approaches to this problem have taken a brute force
approach, and produced a "probabilistic" GRN as their output. However, the
generation of probabilistic edges inadvertently allows spurious
transitions. As a result, our approach models the GRN as a set
of Boolean relations, rather than a single probabilistic GRN.
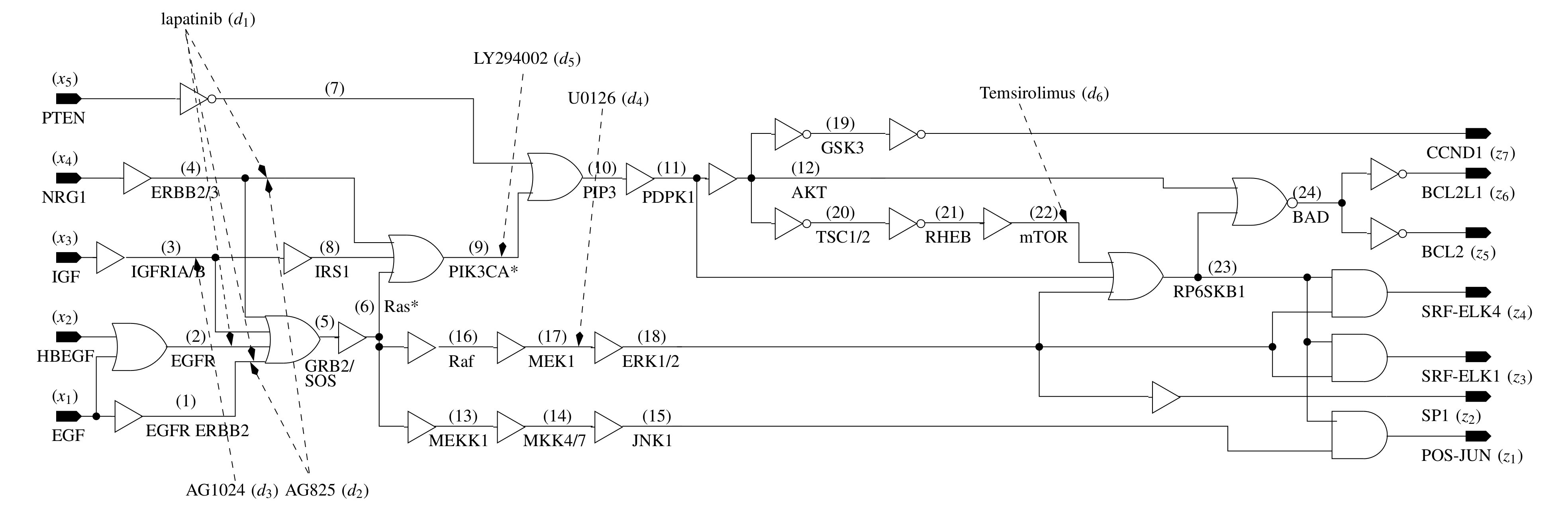
We have also developed techniques to find the least cost drug regime for
cancer, which yields the "best" cure. This is based on a partial max-SAT
framework, using concepts from VLSI testing. Our formulation is general,
implicit and extremely fast.
Other ongoing work in this space is to model the GRN using
"analog" gates, and also to find the logic of each of the nodes in the GRN,
given its structure.
Publications, patents and artefacts:
- "Application of Logic Synthesis to the Understanding and Cure of Genetic
Diseases". Lin, Khatri. Invited paper for special session on non-traditional applications
of CAD algorithms, IEEE/ACM Design Automation Conference (DAC) 2012. To
appear. In this paper, we will discuss our work on predictor inference,
GRN inference, and targeted cancer drug delivery.
- "Efficient Cancer Therapy using Boolean Networks and Max-SAT-based ATPG",
Lin, Khatri. 2010 IEEE International Workshop on Genomic Signal
Processing and Statistics (GENSIPS). Dec 2011, San Antonio, TX. This
paper reports our work on a partial max-SAT based approach to find
minimum number (or cost) of drugs that can force the GRN into a state
which is closest (in a Hamming distance sense) to a normally functioning
individual's GRN.
- "Inference of Gene Predictor Set using Boolean
Satisfiability", Lin,
Khatri. 2010 IEEE International Workshop on Genomic Signal Processing and
Statistics (GENSIPS). Nov 2010, Cold Spring Harbor, NY. pp 1-4. In this
paper, we utilize logic synthesis ideas to find all compatible sets of
genes (predictors) that can influence the behavior of a particular gene
in the state machine. From this information, we perform a back-end
statistical analysis to provide the most likely predictor for each gene
in the GRN
- "Using Boolean Networks and Max-SAT-based ATPG for Efficient Cancer
Therapy". Lin, Khatri. Invited submission for special issue of BMC
Genomics. This paper is an extended version of our GENSIPS-11 paper.
- "Logic Synthesis applied to Genetic Disease Modeling and Cure", Lin,
Khatri. Contract for this brief research monograph submitted to Springer
Publishers. The brief is expected to be published in 2012.

